-
AuthorPosts
-
06/06/2017 at 10:12 AM #2714
tsieger
ParticipantHi,
please, is it possible to read the coordinates of the reconstructed stimulation contacts from some.matfile in the patient directory?I noticed that there is
ea_reconstruction.matfile in the patient directory, containing several different sets of coordinates,
perhaps 2×4 contacts in AC/PC, native, MNI, and subcortically-refined (i.e. brainshift-corrected) spaces:a=load('ea_reconstruction.mat); >> a.reco.acpc.coords_mm{1} ans = 12.9738 -0.5765 -6.4287 13.2689 0.2484 -4.6312 13.5639 1.0734 -2.8338 13.8590 1.8984 -1.0363 >> a.reco.native.coords_mm{1} ans = 9.9016 19.6273 -73.0613 10.4545 20.1438 -71.2103 11.0074 20.6603 -69.3593 11.5604 21.1769 -67.5083 >> a.reco.mni.coords_mm{1} ans = 14.9746 -10.1242 -15.7690 15.5192 -9.0578 -13.6511 15.9094 -8.0424 -11.3782 16.1868 -7.0526 -9.0660 >> a.reco.scrf.coords_mm{1} ans = 10.3017 24.3384 -75.7406 10.8587 24.8301 -73.8912 11.4157 25.3217 -72.0418 11.9727 25.8134 -70.1924Please, what do the individual coordinates refer to?
Are they corrected for brainshift?(It seems that the subcortically-refined coordinates are native coordinates corrected for brainshift, not MNI coordinates. Are the MNI coordinates also brainshift-corrected, or not? If not, can one easily compute brainshift-corrected MNI coordinates?)
Thanks.
Best,
Tomas06/06/2017 at 12:46 PM #2717andreashorn
KeymasterHi Tomas,
– the mni is in mni space and brainshift corrected (if you did perform brainshift correction).
– native is in space of the postop image (e.g. postop_tra.nii or rpostop_ct.nii)
– scrf is in space of the anatomical images (e.g. anat_t1.nii) and is similar to native but brainshift corrected – i.e. if you compare the two, you should see the amount of brainshift
– the acpc fields are coordinates relative to the AC/PC system (auto-converted/marked as defined in Horn 2017 neuroimage).Also thanks for your nice comments in the other post from my side :)
Best, Andy
06/06/2017 at 5:11 PM #2721tsieger
ParticipantHi Andy,
thanks for your comprehensive answer.
So, comparing scrf and native coordinates, I can estimate the amount of brainshift, e.g. more than 4 mm in the antero-posterior direction in one patient:>> a.reco.scrf.coords_mm{1}(1,:)-a.reco.native.coords_mm{1}(1,:)
ans =
-1.2301 4.3909 0.4404Let me please ask one mow question. I’m curious why, if I apply the brainshift, I usually (in most patients) get electrodes missing the STN (fig. below, A), while when omitting the brainshift correction, the electrodes go thru STN, as expected (fig. B). The weird thing is that the brainshift correction seems to make sense for this patient (C), so it is not clear to me why it failed to make the position of the electrodes more accurate. Perhaps there is a problem with the normalization of the post-operative MRI (D)? Apparently, the post-operative MRI is not aligned with the MNI template well enough (D), while the pre-operative is (C).
Could you please comment on this? What would be the remedy for this situation? I would love to correct for the apparent brain shift, but the correction seems to make things worse.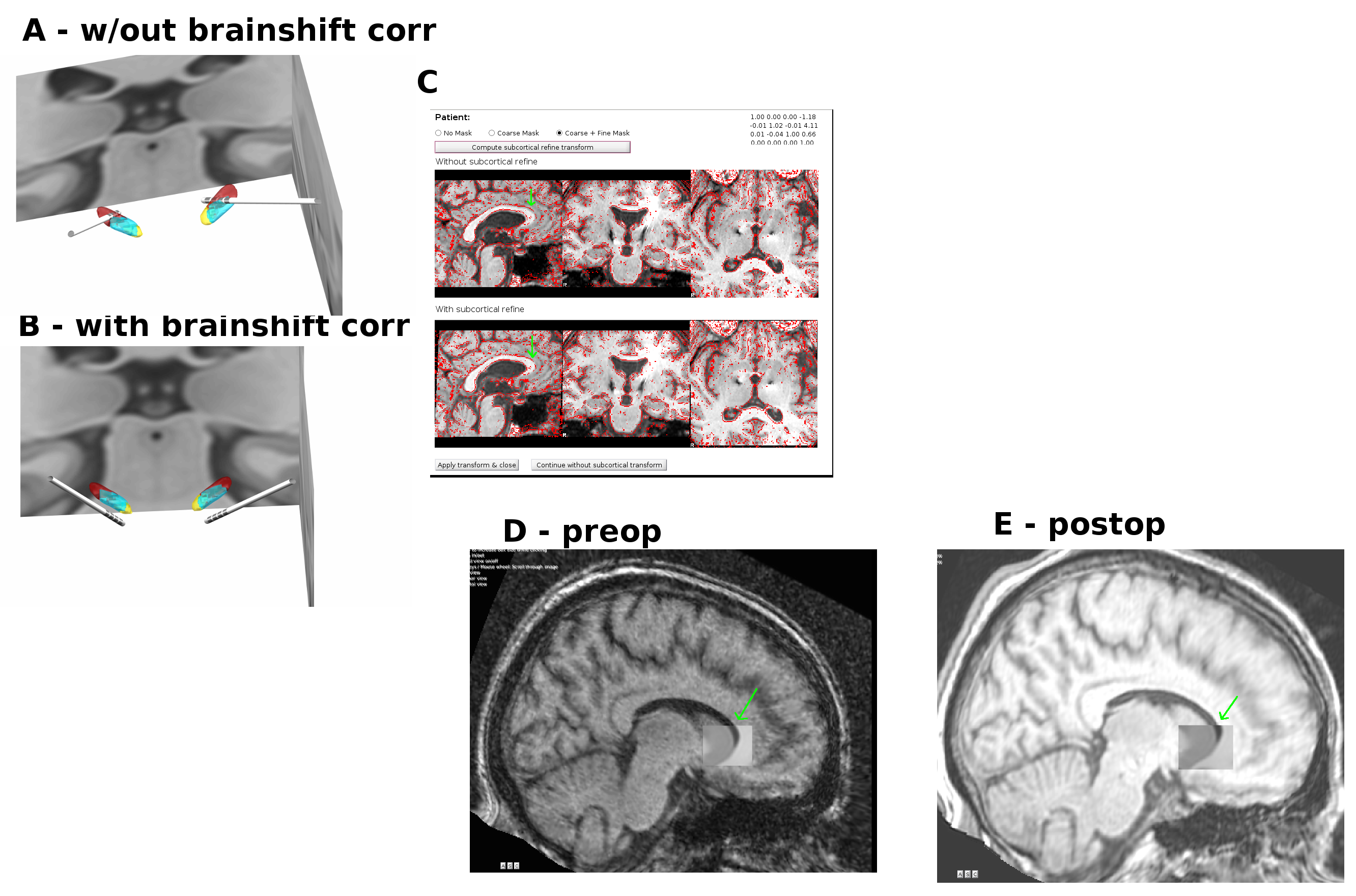
Thanks.
Best, Tomas
06/07/2017 at 9:10 AM #2725tsieger
ParticipantOops, I accidentally swapped some pictures in the figure above, and made the text of my previous post #2721 confusing. I’m very sorry for that.
The figure above is OK now, and the corrected problem description is:
If I apply the brainshift correction, I usually (in most patients) get electrodes missing the STN (fig. B), while when omitting the brainshift correction, the electrodes go through STN, as expected (A). The weird thing is that the brainshift correction seems to make sense for this patient (C, green arrows), so it is not clear why the correction moved the electrodes away from the STN. Perhaps there is a problem with the MRI normalization? Apparently, the post-operative MRI is not aligned with the MNI template well enough (E), while the pre-operative is (D).
What would be the recommended way of dealing with this problem? Shall I perform another MRI normalization? Shall I omit the brainshift correction? (Of course, I would like to have the position of my electrodes reconstructed as accurate as possible, so I would like to perform the correction.)Thanks, Tomas
06/09/2017 at 4:52 AM #2732andreashorn
KeymasterDear Tomas,
this indeed looks a bit worrisome and I’d love to inspect that subject myself. Could you please send it to me in anonymized form? Best would be the complete processed folder with all the files, if possible.
We have had trouble with brainshift correction in the past and fixed it recently. But I assume you use the newest version and since you even have applied the hotfix, this shouldn’t be the cause.
Thanks so much, best, Andy
06/12/2017 at 2:33 PM #2742tsieger
ParticipantDear Andy,
sure, I’m sending a link to the patient folder data directly via email.Yes, I’m using the latest version of LeadDBS 1.6.3.4, with the hotfix from 2017-06-12 morning applied.
Best, Tomas
06/16/2017 at 6:03 AM #2775andreashorn
KeymasterSo just in case someone else reads this, the issue could be resolved and was due to a misregistration when applying the coarse+fine mask in the brainshift removal tool on tsieger’s patient.
-
AuthorPosts
- The forum ‘Support Forum (ARCHIVED – Please use Slack Channel instead)’ is closed to new topics and replies.

